ReactionString/RestScan
These features were developed by the Preferred Networks Research Team (the Matlantis development team) and implemented in Matlantis. This page provides an overview of these powerful tools.
ReactionString
ReactionString is a reaction path search algorithm similar to NEB, Dimer, and Climbing NEB. Unlike conventional methods, it automatically adjusts the number of intermediate structures ("images") along the reaction path, densely sampling near the transition state (TS). This results in TS accuracy that is comparable to the Dimer method, and generally more precise than NEB. ReactionString also handles multiple local minima without issue and simultaneously optimizes both the reaction path and TS, helping avoid the kind of failures sometimes encountered in IRC (Intrinsic Reaction Coordinate) calculations—such as ending up at an unexpected structure. Thanks to these features, ReactionString can explore complex reaction coordinates involving multiple TSs between the initial and final states (as illustrated below). One potential drawback is that it can take longer to compute than NEB, but options such as early termination based on runtime are available to manage this.
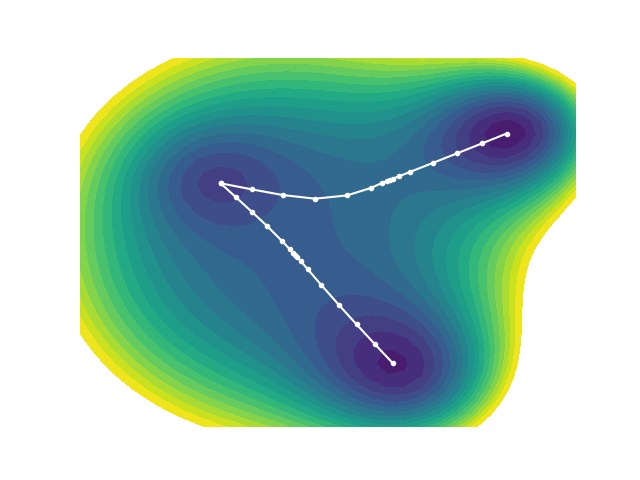
Below is an example of the synthesis of methyl methacrylate (MMA) over a palladium (Pd) catalyst.
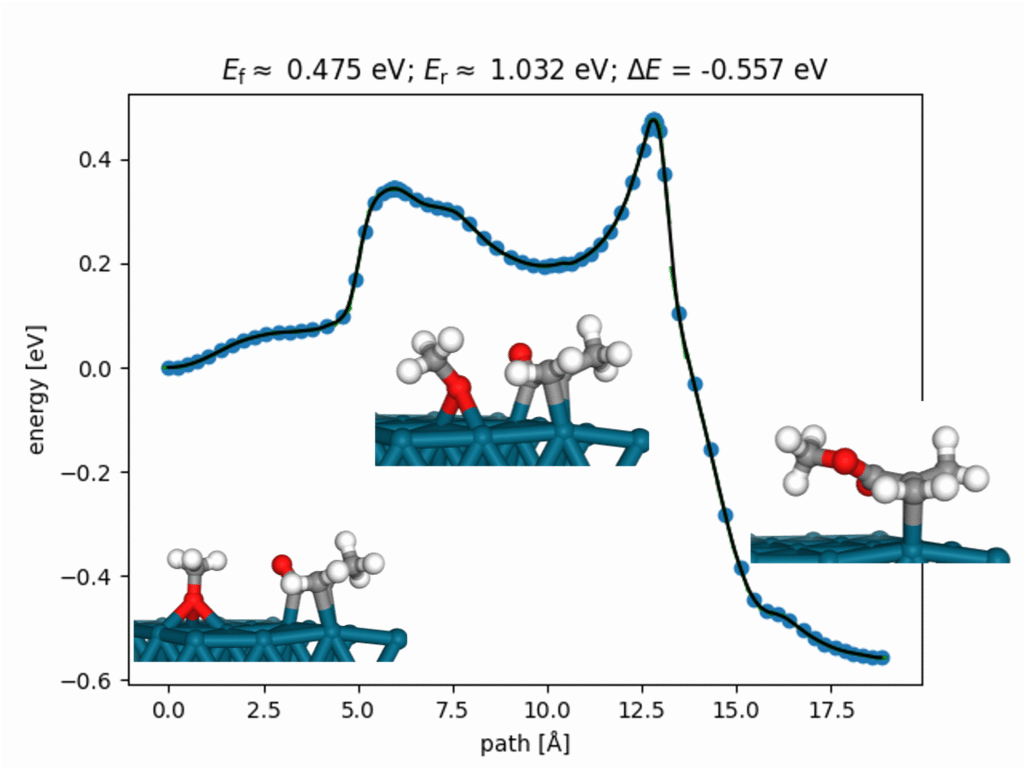
RestScan
RestScan stands for restraint scan. It allows part of a molecule’s geometry to be constrained and continuously modified until a specified condition is met—including inequality conditions. This enables sequential processing even with an imprecise understanding of the reaction coordinate. RestScan is particularly useful for preparing initial or final structures required in reaction path searches such as ReactionString or NEB.
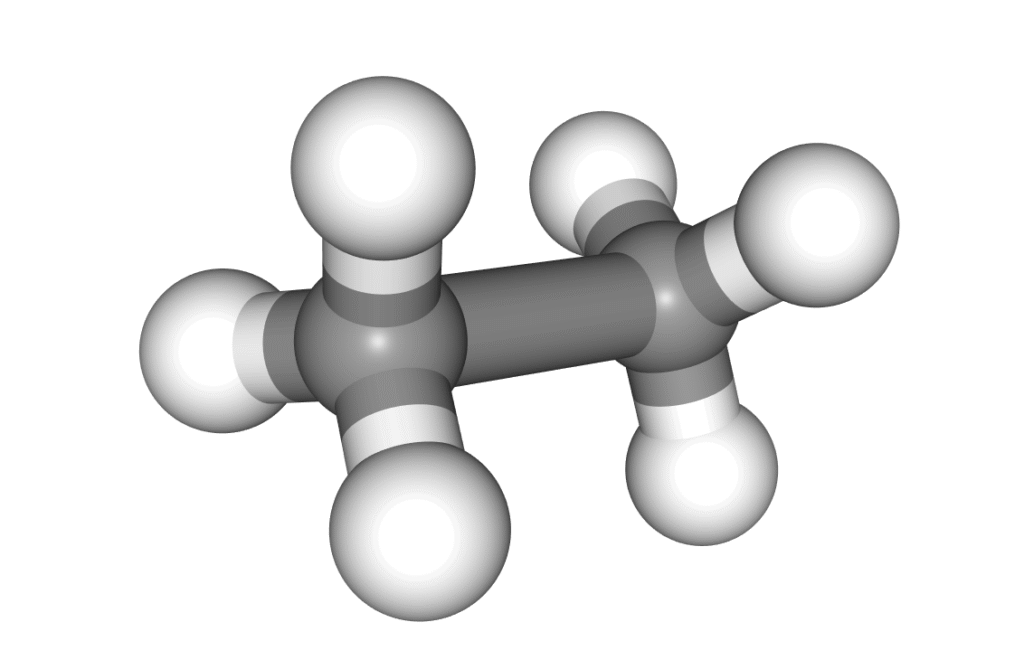
In the example below, a methane molecule is rotated 360 degrees around a C–C bond, and the scan is set to stop once it passes an energy barrier.
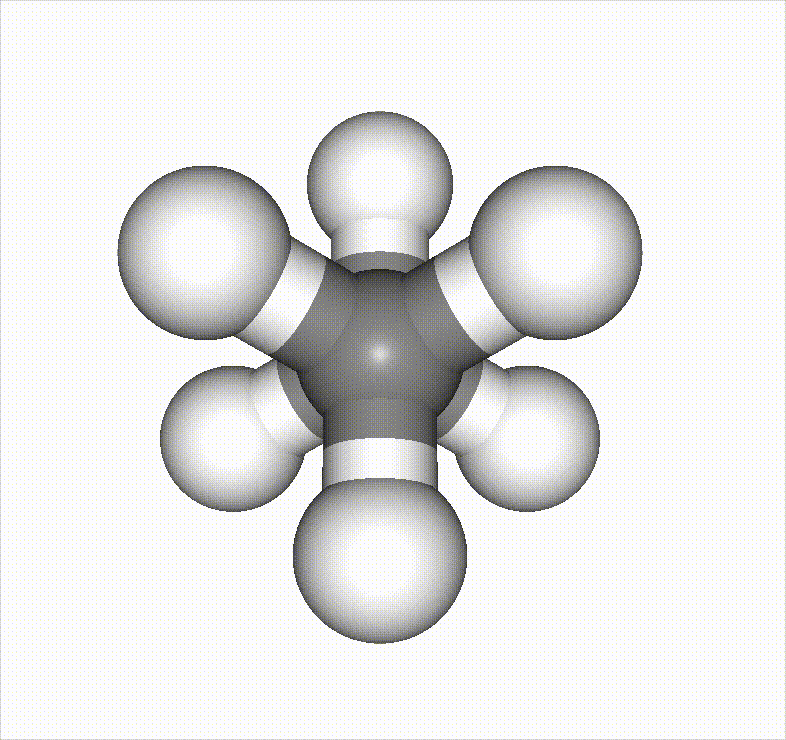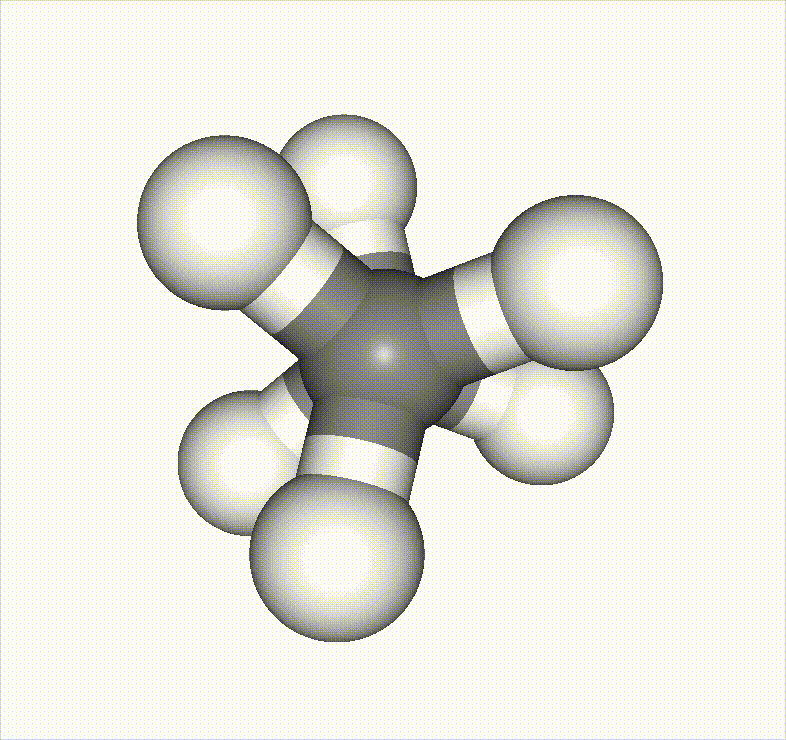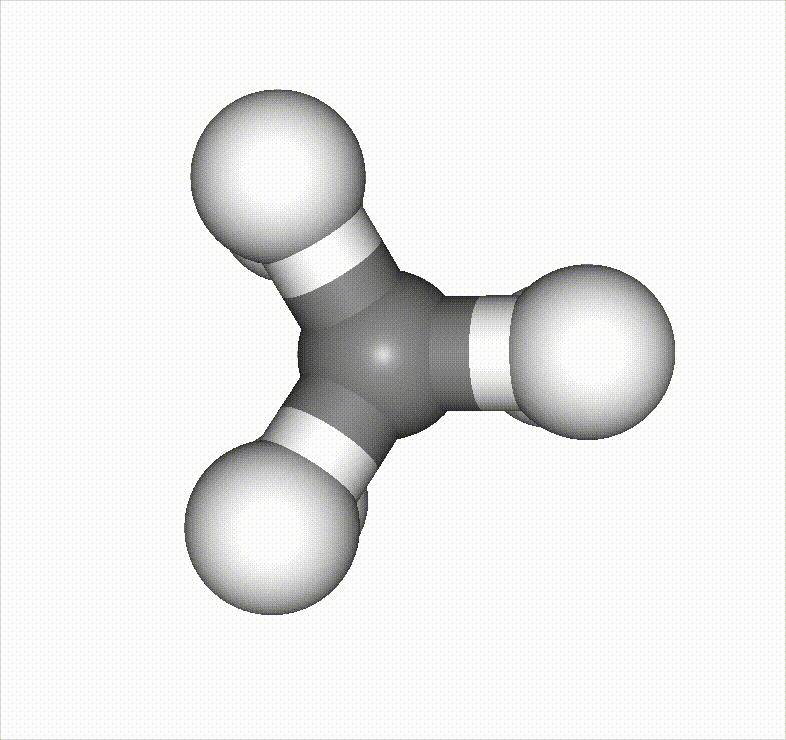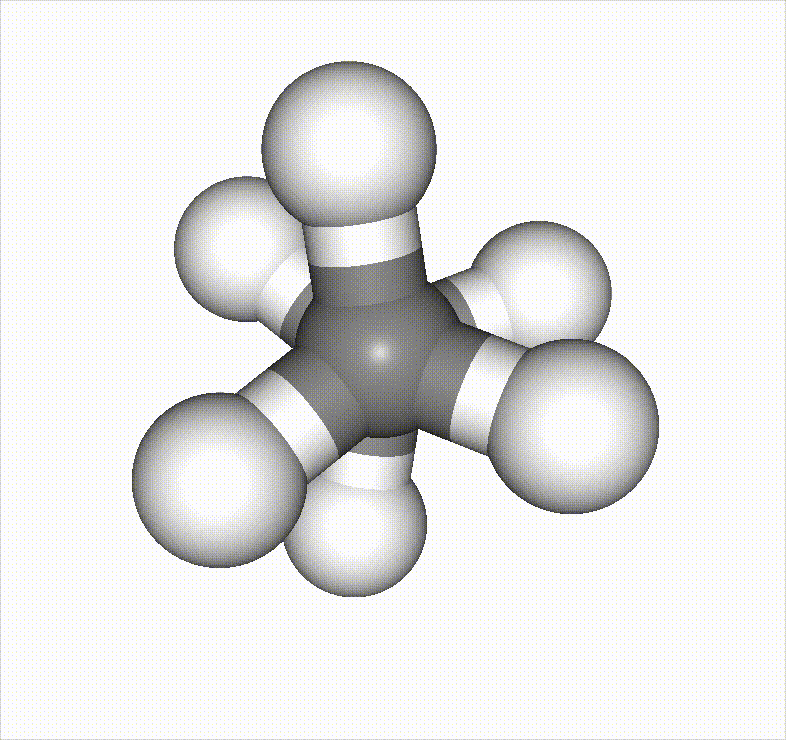
Reference material: https://speakerdeck.com/pfn/matlantis-user-conference-3-hayashi
A paper detailing the methodology is currently in preparation.